Premashis Manna
Contact Information
Assistant Professor
Areas of Expertise
- Analytical
- Physical
Bio:
Premashis (Prem) Manna earned his B.Sc. and M.Sc. in Chemistry from Ramakrishna Mission Vidyamandira, Belur (Calcutta University) and the Indian Institute of Technology Kanpur (IIT Kanpur) in 2010 and 2012, respectively. He completed his Ph.D. in Chemistry at the University of Colorado Boulder, working under the guidance of Dr. Ralph Jimenez. His doctoral research focused on using high-throughput microfluidic platforms to engineer brighter red fluorescent proteins for bioimaging and biosensing applications. Additionally, he investigated the photodynamics of these proteins, particularly their blinking and photo-switching behaviors. After completing his Ph.D. in 2017, Dr. Manna joined Prof. Gabriela Schlau-Cohen's lab at MIT as a postdoctoral researcher. There, he studied light-harvesting complexes at the single-molecule level, developing a method to quantify protein-protein interaction strengths of membrane proteins under various conditions using single-molecule data and statistical mechanical principles. In late 2022, Dr. Manna joined Gilad Haran's lab at the Weizmann Institute of Science, where he leveraged single-molecule FRET under plasmonic environments to enhance the time resolution of such measurements. Dr. Manna began his role as an assistant professor at the Department of Chemistry and Biochemistry, The Ohio State University (OSU) in January 2025. At OSU, his lab integrates high-throughput microfluidic-based selection methods with single-molecule techniques to study and engineer photo-switching proteins, light-harvesting complexes, and plastic-degrading enzymes.
Research Overview:
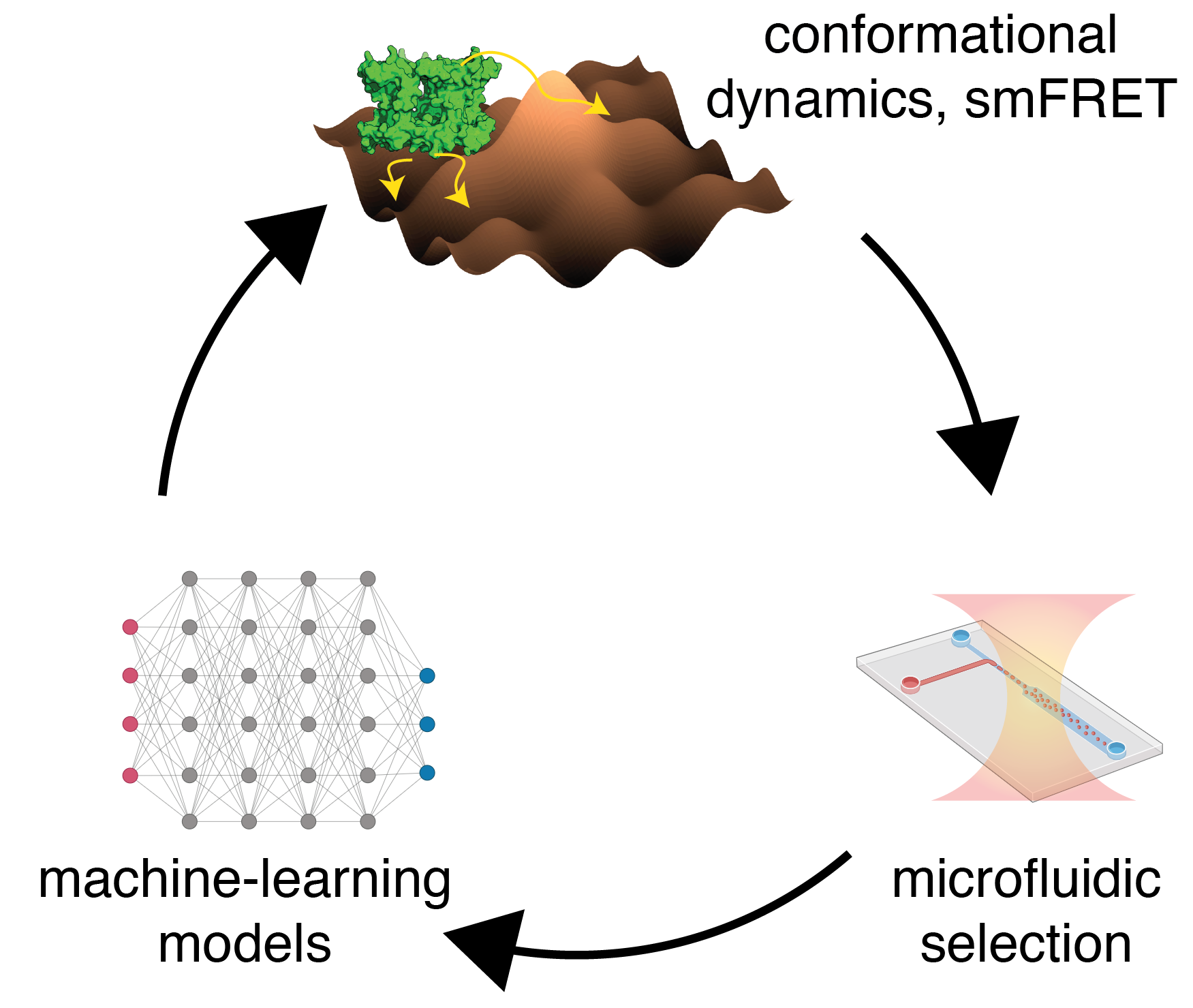
Dynamics-guided directed evolution of protein function.
Directed evolution has proved to be a powerful technique to alter the protein functions for their industrial and therapeutic applications via genetic diversity and selections thereafter. So far, this technique has been employed based on the read-outs from equilibrium properties of proteins such as fluorescence, pH stability, thermostability, etc. By employing machine-learning models and the spectroscopic read-outs from protein conformational fluctuations, my laboratory will develop a dynamics-guided directed evolution platform. This platform will help us to develop efficient enzymes, novel antenna complexes, and bio-markers as discussed below.
Biosensors for single-molecule and super-resolution microscopies. Fluorescent proteins (FPs)-based biomarkers have remained state-of-the-art in biological assays as they can be expressed on proteins of interest at stoichiometric levels. In the last decade, directed evolution has achieved a remarkable feat in enhancing the brightness of these FPs. However, the low photostability and high blinking make these probes unsuitable in single-molecule applications where high illumination intensities are used. On the other hand, localization-based super-resolution techniques like PALM or STED rely on the on/off switching of the fluorophores. Our laboratory aims to develop bright photoswitching FPs for advanced nanoscopy applications like MINFLUX by controlling their dark state conversion (DSC) and ground state recovery (GSR) processes which ultimately relates to their blinking properties and photostability.
Far-red absorbing light-harvesting complexes. A promising avenue to enhance photosynthetic efficiency in plants is to extend the photosynthetically active radiations (PAR) to the far-red. Green plants use only the blue and red light for oxygenic photosynthesis leaving the green and infrared parts mostly unused. It can be shown that a modest extension in the PAR to 750 nm could lead to a 20% increase in photosynthetic efficiency. By employing a cohort of biochemical, biophysical, and bioinformatics tools, our laboratory aims to optimize the pigment-protein interactions beneficial for further far-red absorbing plant species.
Enzymes for fast and efficient plastic degradation. Polyethylene terephthalate (PET), a semi-aromatic polyester, is one of the highly-consumed plastics with high resistance to biodegradation. However, recently discovered PET-degrading enzymes such as PETase or leaf-branch cutinase (LCC) offer promise to depolymerize PET in an environmentally friendly manner. First, our laboratory will investigate the mechanistic aspect of such plastic-degrading enzymes using various bulk and single-molecule techniques. Later, insight gained from such studies will be employed to generate enzymes capable of degrading other kinds of plastic such as polyethylene via directed evolution approaches.
Publications:
Premashis Manna*, Madeline Hoffmann, Thomas Davies, Matthew P. Johnson, and Gabriela S. Schlau-Cohen, “Energetic driving force for LHCII clustering in plant membranes”, Science Advances, 2023, 9, eadj0807 (*joint first and corresponding author).
Sriit Mukherjee, Premashis Manna*, Sheng-Ting Hung, Felix Vietmeyer, Pia Friis, Amy Palmer, and Ralph Jimenez, “Directed evolution of a bright variant of mCherry: Suppression of non-radiative decay by fluorescence lifetime selections”, The Journal of Physical Chemistry B, 2022, 126, 4659-4668 (* joint first and joint corresponding author).
Premashis Manna, Thomas Davies, Madeline Hoffmann, Matthew P. Johnson, and Gabriela S. Schlau-Cohen, “Membrane-dependent heterogeneity of LHCII characterized using single-molecule spectroscopy”, Biophysical Journal, 2021, 120, 3091- 3102.
Premashis Manna, Sheng-Ting Hung, Srijit Mukherjee, Pia Friis, David Simpson, Maria Lo, Amy Palmer and Ralph Jimenez, “Directed Evolution of Excited State Lifetime and Brightness in FusionRed using a Microfluidic Sorter”, Integrative Biology, 2018, 10, 516-526.
